Molecular Basis of Inactive B-RAFWT and B-RAFV600E Ligand Inhibition, Selectivity and Conformational Stability: An in Silico Study
Filip Fratev *†, Svava Ósk Jónsdóttir †, Elina Mihaylova ‡ and Ilza Pajeva §
Center for Biological Sequence Analysis, Department of Systems Biology,
Technical University of Denmark, Kemitorvet, Building 208, DK-2800 Kongens Lyngby, Denmark,
Micar Ltd., 39 Asparuh Str., 1000 Sofia, Bulgaria, and
Centre of Biochemical Engineering “Ivan Daskalov”, Bl. 105 Acad G. Bontchev Str., 1113 Sofia, Bulgaria
Mol. Pharmaceutics, 2009, 6 (1), pp 144–157
DOI: 10.1021/mp8001107
Publication Date (Web): December 2, 2008
Copyright © 2008 American Chemical Society
Section:Enzymes
Abstract
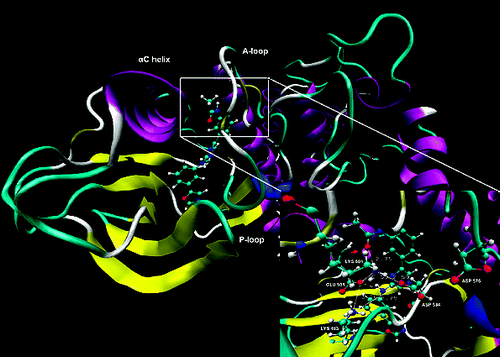
The B-RAF kinase plays an important role both in tumor induction and maintenance in several cancers. The molecular basis of the inactive B-RAFWT and B-RAFV600E inhibition and selectivity of a series of inhibitors was examined with a combination of molecular dynamics (MD), free energy MM-PBSA and local-binding energy (LBE) approaches. The conformational stability of the unbounded kinases and in particular the processes of the B-RAFV600E mutant activation were analyzed. A unique salt bridge network formed mainly by the catalytic residues was identified in the unbounded B-RAFs. The reorganization of this network and the restriction of the active segment flexibility upon ligand binding inhibit both B-RAFWT and B-RAFV600E, thus appearing as an important factor for ligand selectivity. A significant correlation between the binding energies of the compounds in B-RAFWT and their inhibition effects on B-RAFV600E was revealed, which can explain the low mutant selectivity observed for numerous inhibitors. Our results suggest that the interactions between the activation segment and the αC-helix, as well as between the residues in the salt bridge network, are the major mechanism of the B-RAFV600E activation. Overall data revealed the important role of Lys601 for ligand activity, selectivity and protein stabilization, proposing an explanation of the observed strong kinase activation in the K601E mutated form.
